
The Key Benefits of MMS
Microfluidic Modulation Spectroscopy (MMS) is a novel and advanced technique for biomolecular characterization that offers several advantages compared to traditional IR-based protein characterization methods. Some of the key benefits of MMS are:
- High Sensitivity: MMS utilizes a tunable Quantum Cascade Laser (QCL) that is at least 1,000 times more intense than conventional light sources, allowing for a maximal signal-to-noise ratio to achieve ultra-high sensitivity. This sensitivity allows MMS to see changes missed by other technologies, giving you greater confidence in your biophysical characterization, and saving time and money.
- Automation: MMS can analyze multiple samples with hands free operation, making it a highly efficient and cost-effective solution for large-scale biomolecular characterization.
- Minimal Sample Preparation: Unlike other techniques that require extensive sample preparation, MMS is a more user-friendly solution for biomolecular characterization and monitoring changes in protein structure and stability.
- Label-free: MMS is a label-free technique, meaning that it does not require the use of fluorescent dyes or other exogenous labeling agents, making it a safer and more cost-effective solution for biomolecular characterization. Allowing the measurements to be performed in formulation buffer at formulation concentration for more realistic and relevant structural information.
- Non-destructive protein analysis: MMS is a non-destructive technique, and the integrated software allows for simple sample recollection post-analysis. MMS has demonstrated a recovery rate of 95% and a >99% structural similarity relative to the starting material, making MMS a fantastic solution for samples that are limited in availability or volume.
Overall, MMS provides a unique and innovative solution for biomolecule structural analysis and biomolecule detection, offering a high degree of accuracy, sensitivity, and repeatability compared to other methods.

Learn how you can save time and money in your biotherapeutic development process. Access MMS data now!
What is Microfluidic Modulation Spectroscopy technology?
MMS is an ideal protein characterization tool for measuring changes to higher order structure (HOS) of biomolecules and can be applied to many fields utilizing therapeutic proteins, such as QA in biologics development.
- 20x faster and 30x more sensitive to changes in structure than CD or FTIR* to help you flag unsuitable candidates earlier and meet or beat your project deadlines
- Novel, fully automated technique for higher-order structural measurements of proteins and biomolecules offering walk-away convenience and simple platform methods, even for researchers without expertise in spectroscopy
- Accurate and reproducible measurements with a broad concentration range from 0.1 mg/mL to >200 mg/mL allowing measurements in native conditions and the formulation buffer of interest
- Real-time buffer subtraction minimizes background noise and interference from formulation excipients, resulting in a significant advantage in data accuracy and utility over CD and FTIR
*Journal of Pharmaceutical Sciences, JAN 2020
Why Add MMS Into Your Biotherapeutic Development Workflow?
- DISCOVERY: Integrate structural monitoring to add more robust selection and discrimination criteria for candidate screening
- FORMULATION: Automatically analyze and compare samples across a formulation study to predictively identify optimal buffer formulations, stability profiles, and ideal storage conditions earlier in the process to eliminate costly downstream failures.
- MANUFACTURING: Track stability and structure as Critical Quality Attributes (CQAs) across the entire manufacturing process to maximize safety, efficacy, and functionality
How MMS Adds Value to Your Biotherapeutic Development Toolkit
By incorporating MMS into your analytical suite of tools, you will increase the efficiency and utility of your characterization process by monitoring stability, structure, similarity, and intermolecular aggregation all from a single automated run. With the benefits of MMS plus our state-of-the-art intuitive analytical engine, delta software, you'll gain the confidence in your data that no other biophysical characterization platform can provide.
Learn more about MMS in our newly published app note on the rapid and accurate detection of the higher-order structure of a protein library using MMS!
Frequently Asked Questions
What are some benefits of measuring secondary structure compared with the other levels of protein structure?
Secondary structure provides direct access to the protein’s function and stability since the global protein structure and shape are closely related to the relative abundance of secondary structural motifs. Changes in the global properties of the protein are often accompanied by secondary structural change. Compared to tertiary and quaternary protein structures which require high experimental effort, secondary protein structure is comparably easy to access, for example through vibrational spectroscopy. On the other hand, secondary protein structure also serves as an ideal reporter on local structural changes, e.g. through binding events or the onset of protein structural aggregation processes.
Are bioinformatics used to measure change in protein structure?
Bioinformatics are most often linked to the organism genome and protein primary sequence as opposed to the structure. However, these tools can absolutely be useful in combination with protein structure characterization to help complete the story. For example, the figure below shows how it is possible to link up the primary sequence to the three-dimensional structure. Bioinformatics could help identify evolutionarily conserved regions, like active sites or binding pockets of the protein and therefore, regions of most importance for the protein function.
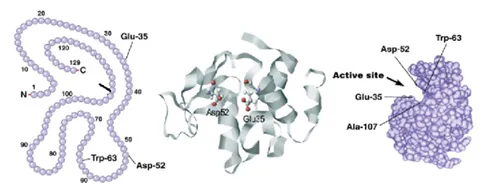
How does protein sequencing differ from measurement of protein higher order structure?
Protein sequencing provides the sequence of amino acids within one protein while measurements of protein higher order structure provide the protein’s 3D arrangement either locally (secondary) or globally (tertiary and quaternary).


