
Protein Stability
Why Protein Stability Matters in Biotherapeutic Development
Stability may be considered one of the most important structural characteristics to monitor during development and formulation since it directly relates to the function of a biotherapeutic. When the secondary conformation of a protein changes, the active sites could be affected, oftentimes detrimentally. Using the Aurora TX to monitor the stability of a protein over an experimental set of conditions is simple and intuitive. By tracking specific wave numbers that are associated with characteristic secondary structural features and calculating a “similarity score” or using weighted spectral difference, it is possible to observe if a protein changes its conformation which may indicate degradation, denaturation, or possibly aggregation.
How MMS Technology Tracks Protein Stability
Analyzing samples using Microfluidic Modulation Spectroscopy (MMS) provides a level of sensitivity that is unrivaled. Enhanced second derivative results are generated from the Absolute Absorbance spectra and further converted to a Delta plot highlighting subtle changes in the secondary structure. This enables early action to inform decisions for upstream development modifications and downstream formulation workflow optimization.
Stability Analysis Example: BSA Sample Comparison

Delta plot and stability analysis of 13 commercially sourced BSA samples compared to the control. Second derivative spectra for each biosimilar were subtracted from ref sample 4112305 to highlight spectra differences. Stability analysis tracks signals corresponding to anti-parallel beta-sheet, parallel beta-sheet, and alpha-helix for each sample. Application Note Biosimilar Structural comparison of Commercially Source Reference Standards by MMS Rapidly Detects Subtle but Critical Differences to Correctly Predict Activity for Use in an ELISA Product.

FAQs About Protein Stability and Detection
Is it possible to increase protein solubility using formulations to prevent precipitation and protein aggregation?
Formulations can absolutely help increase solubility! One of the most important features to get right in a formulation buffer is the pH. In addition to finding the right buffer, there are also many different additives that can help stabilize or solubilize your protein. Some of these additives include salts to help balance ionic strength, sugars to help with the protein’s hydration, detergents to act as an emulsifier or to protect the protein from aggregating, and amino acids can help balance charges.
What does the term biomolecular stability refer to?
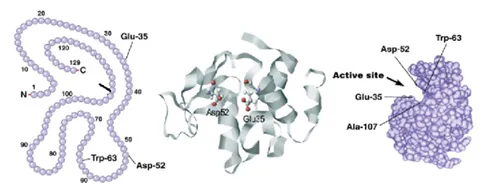
Biomolecular stability refers to the ability of a biomolecule to withstand external stress, i.e., to keep its designated structure and activity. Most often increasing temperature is used to determine stability, but other stresses could be used too, such as pH or agitation. The relevant stability is determined by the stress-induced loss in a structure that already gives rise to a loss of function, too. This could be due to a structural change around the active site of that protein. In addition to that, proteins often have intermediate states that still conserve some part of their native structures and with some residual stability. The figure below shows these different states as an example of a thermally stressed protein (apoflavodoxin).

Which molecules are most susceptible to changes in stability?
In general, protein molecules that are neutral in surface charge or have hydrophobic surfaces are prone to interact with each other and hence cause changes in stability. This propensity is highly dependent on the primary amino acid sequence as well as which types of amino acids are dominant in the protein. If there are more hydrophilic and charged amino acids on the surface of the proteins, they will be less susceptible to aggregation caused by surface interactions. On the other hand, if the proteins are folded in such a way that a lot of the hydrophobic amino acids are exposed on the surface, they will be more susceptible to changes in stability. As a result, proteins that have mutations in their primary sequence are likely to suffer from stability issues. In specific applications, however, site-specific mutations are designed on purpose to enhance the stability or activity of a protein.
What causes proteins to lose stability?
Unavoidable changes in temperature, pH or just moving the sample can trigger, for example, oligomerization or unfolding processes which can decrease the stability of the protein towards further stresses. The mechanisms that lead to protein instability can be diverse. The different mechanisms that can lead to a loss of stability in monoclonal antibodies are summarized in a review by Le Basle et al. (Le Basle, Yoann, et al. Physicochemical stability of monoclonal antibodies: a review." <em>Journal of Pharmaceutical Sciences</em> 109.1 (2020): 169-190.)
Which protein structure is most stable?
Protein stability is dictated by the type of interactions that help the folding of the entire protein molecule. The primary structure of proteins is the most stable since the protein sequence is held together by covalent bonds between individual amino acids. Some of the tertiary structures that are held together by disulfide bonds are the most stable other than the primary sequence of the protein. Among the non-covalently bound structures, protein secondary structure is the most stable due to the predominant hydrogen bonding interactions.
Can loss of stability occur at all steps of the drug development process?
Yes, loss of stability can occur at all steps of the drug development process, including early discovery, pre-clinical research and development, formulation, and clinical development. Common factors that can cause the loss of protein stability in early discovery stage include process temperature of proteins, freeze/thaw cycles of proteins, agitation stress of proteins, protein concentration, etc. In the later drug development stage, such as formulation, the goal is to maximize protein stability by choosing the best excipients and buffers for the proteins.
How is stability detected and measured (tools, etc)?
Stability can be defined in many ways based on what the applied stress is. In most cases, stability is inferred based on the protein’s melting temperature (Tm). If a protein can withstand higher temperatures without unfolding, that protein is said to be more stable. There are multiple tools for measuring melting temperature including DSF, DSC, and CD.
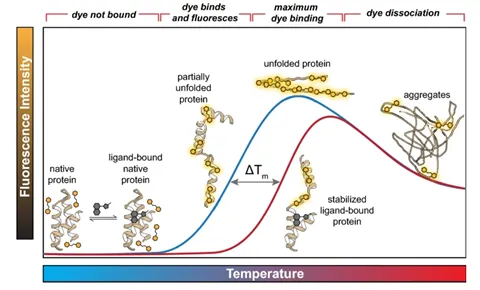
The figure above shows the quantification of stability through the melting temperature determined by differential scanning fluorimetry (DSF). The melting temperature is the temperature that causes the protein to be halfway unfolded, and the higher the melting temperature, the more stable the protein. This figure shows that the melting temperature increases when it’s bound to a ligand. (Samuel, Errol LG, et al.) "Processing binding data using an open-source workflow." Journal of Cheminformatics 13.1 (2021): 1-11.)
However, there are many other forms of stress other than temperature such as agitation, pH, pressure, light exposure etc. It is also important to understand the structure and stability of a biologic under all likely production conditions. MMS is another tool that can measure the protein structure after being exposed to any of these stresses and that can help predict the protein stability.
Which secondary structures are associated with stability loss and the current limitations to detect such?
The loss of alpha-helix or beta-sheet structure and the formation of intermolecular beta-sheet is associated with stability loss. FT-IR and CD are the most prevalent techniques to detect these secondary structures. However, FT-IR suffers from low sensitivity and reproducibility to detect small changes in the secondary structure. CD on the other hand is limited by a low concentration range and less sensitive towards the prediction of beta-sheet compared to alpha-helix structures.
What can help us prevent loss of stability?
Minimizing the amount of stress that a protein or biologic undergoes (temperature, agitation, pH swings) will help mitigate loss of stability. Additionally, certain excipients can be added to formulation buffers to help protect against the stresses that are unavoidable. Some examples of excipients used to increase stability are amino acids, sugars, salts, or small amounts of surfactants.
The application note titled “Detecting Protein Conformational Change Due to Ligand Binding and Stabilization Using MMS” details the stabilizing effects ligands can have. In this study, a protein was exposed to 50°C overnight. There were 3 different conditions: 1) protein alone(Apo), 2) protein + ligand 1, and 3) protein + ligand 2. The results show that when the protein is exposed to heat stress, there is a large increase in the the beta-sheet features and a decrease in alpha-helix. However, when in the presence of ligand 2 the effect of heat is much less dramatic, and the heat has the least impact on the sample bound to ligand 1. This indicates that ligands 1 and 2 both exhibit stabilizing effects on the protein, and ligand 1 has the most stabilizing effects.
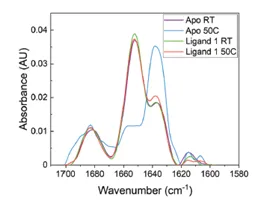

The figures above are from the application note and show MMS data collected of a protein unbound (Apo) and bound to Ligand 1 and how 50°C heat stress overnight affects the structure of the protein. The heat stress drastically changes the structure of the Apo protein but has very little effect on the protein that is bound to ligand 1. This data shows that the ligand stabilizes the protein against heat stress.
Can different formulation buffers be used to increase stability?
Yes! There are many different additives/excipients/preservatives that can be added to formulation buffers to increase a biologic’s stability. It is important that the additives do not interfere with the function or structure of the biologic, so all functional assays and characterization must be completed in the final formulation buffer that includes any additives. Some examples of additives used to increase stability are amino acids, sugars, salts, or small amounts of surfactants.
Batabyal et al. emphasize the importance of proper buffer formulation and characterization in the article “Shaping IR Spectroscopy into a Powerful Tool for Biopharma Characterization.” In this study, polysorbate 80 (PS 80) was introduced to the formulation buffer for a monoclonal antibody. PS 80 is commonly used in aqueous drug products to increase stability; however, it is important that the introduction of PS 80 does not change the structure or function of the drug product. MMS had no issues with buffer or excipient interference and generated clean protein structural information that showed PS 80does not affect the structure of the monoclonal antibody.
A different example can be seen in the application note titled “Using MMS to Measure Buffer-Induced Structural Changes in an Alpha-helix Rich Enzyme,” where the buffering conditions did impact the structure of lysozyme. Four different buffering conditions were tested, and it was determined that lysozyme has the highest fractional contribution of alpha-helical structure when it’s in tris buffer compared to water, phosphate buffer, and PBS. Lysozyme is the most active when in tris buffer and this may be due to the optimized structure with higher alpha-helical content. Since the active site of lysozyme is sandwiched between regions of alpha-helix, it makes sense that loss in alpha-helical content would result in less activity.
Is all stability loss in proteins reversible/irreversible?
Some stability lossesin proteins are reversible but some are irreversible. A method to predict whether the stability loss is reversible or irreversible is to identify what kind of change in the protein is causing instability. If the change comes from the secondary structures, there is a high probability that the stability loss is irreversible, especially in the case of beta-amyloid formation. If the change comes from the tertiary or quaternary structures, the stability loss is more likely to be reversible. For example, reversible aggregation due to the surface interaction of proteins will cause a temporary loss of stability. Reversible stability loss can be fixed by changing the formulation to separate protein monomers.
How do I setup an experiment to find the formulation condition where stability of proteins starts to decline?
Experiments that directly probe the loss in stability would be accelerated stability studies where a protein or biologic is exposed to higher-than-normal stress conditions to help predict when that drug product will fail and gauge the ttheshelf-life and storage conditions. Accelerated stability studies involve testing a range of stresses like temperature, agitation, pH, and light exposure.
An example of an accelerated stability study using MMS can be seen in the application note titled “Biosimilar Comparison and Accelerated Stability Predictions Based on <2% Secondary Structure Differences Using MMS.” In this study, insulin originator (Humalog) and a biosimilar were compared and the biosimilar was exposed to 4 or 30°C for 8 weeks to determine its structural stability. The data show small structural changes due to the heat stress and it is correlated with a loss in alpha-helix, unordered, and turn features, and increase in beta-sheet content. Structural information can be useful for stability studies to understand what happens when a protein becomes unstable.
What can be done if loss of protein stability is detected?
Loss in protein stability can manifest in multiple different ways, I.e., the protein could unfold, self-associate into oligomers, or structurally aggregate into fibrils. The strategy for addressing loss in stability will depend on how the protein is manifesting that loss. For example, colloidal aggregates can be addressed using different formulation conditions to help dissociate the oligomers. However, if the samples are unfolding or structurally aggregating, the solution may be tore-engineer or make mutations to the original construct to make it less hydrophobic.
Why is stability important?
Stability, or the ability of a protein or biologic to withstand stresses, is crucial for its activity in a final formulation. A higher tolerance towards external stresses gives rise to better activity, safety, and a longer shelf-life of the final product.
What are the consequences of loss of stability?
A loss of stability may give rise to a loss in protein activity or may even cause unwanted side effects. A less stable protein is prone to aggregation and irreversible structural changes.

